Current Research Projects
|
Project 1: Mechanisms of Zygotic Genome Activation - A Major Nuclear Reprogramming Event
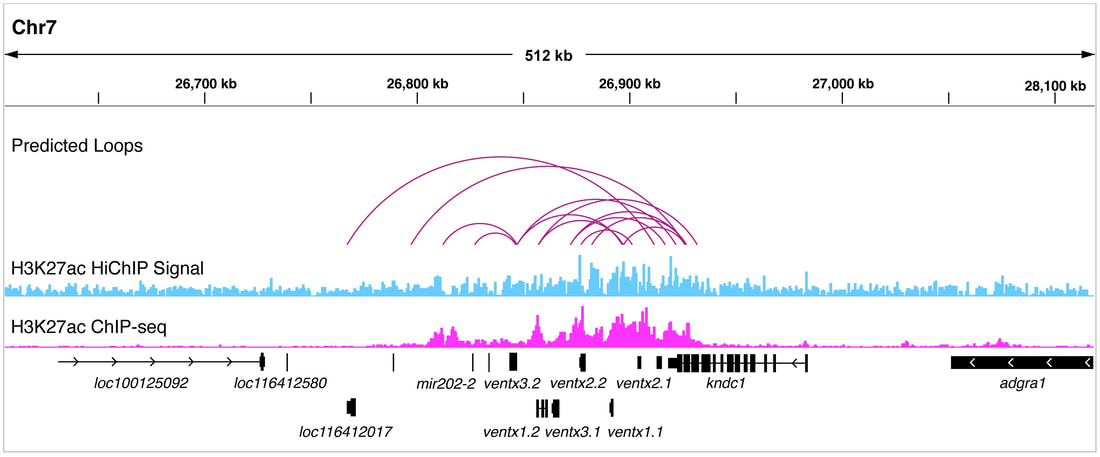
Animal cloning has revealed that differentiated cells can be returned to embryonic states through the transplantation of adult nuclei into eggs. This suggests that the adult cell genome can be reset to an embryonic state within the egg environment, highlighting the presence of maternal factors in eggs capable of reprogramming adult nuclei. We are keen to understand the roles of these maternal transcription factors and the associated chromatin modifiers, which are essential for ensuring precise timing and location of gene expression, crucial for accurate cellular differentiation. Our focus includes the initial stages of genome activation termed zygotic genome activation. We investigate how transcription factors present in eggs influence cell fates and imprint essential epigenetic markers on the embryonic genome. Additionally, our research sheds light on the intricate relationship between chromatin dynamics and the combined actions of maternal and zygotic transcription factors.. |
|
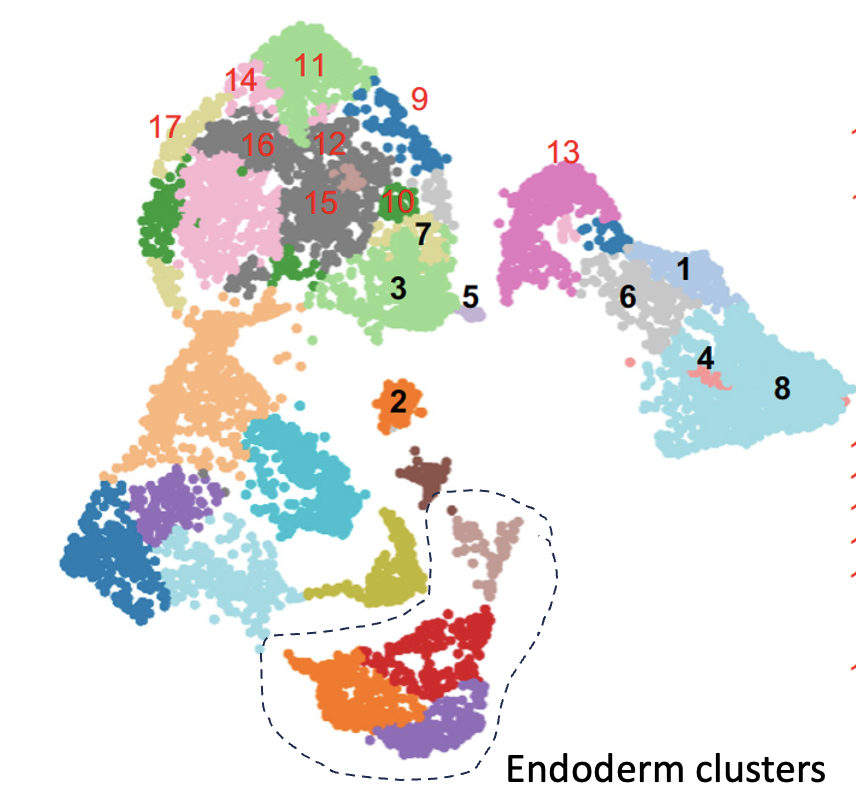
Project 2: Decoding the Regulatory Program of Embryonic Development Embryonic development, a complex journey from zygote to a fully formed organism, is orchestrated largely by cis-regulatory modules (CRMs), often referred to as enhancers. These CRMs are essential, ensuring the precise timing and location of gene expression throughout embryogenesis. Surprisingly, despite the hundreds of thousands estimated to regulate approximately 20,000 genes in vertebrates, the activities of these CRMs in development remain unstudied. Notably, mutations within these CRMs can lead to various genetic disorders and embryonic defects, highlighting the critical need for in-depth research. Our investigations utilize cutting-edge sequencing methods, like single-cell transcriptomics, to probe CRM activity in Xenopus embryos. Given their evolutionary relevance, these embryos provide a unique window into vertebrate gene regulation. Our objective is to decode the secrets of early development and the complex regulatory systems governing embryogenesis. |
|
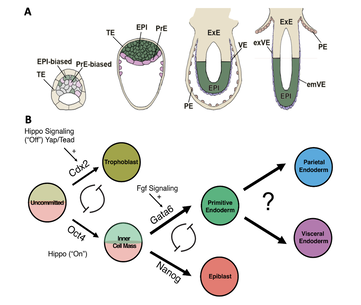
Project 4: Mammalian Extraembryonic Endoderm Development:
Mammalian extraembryonic tissues are pivotal for early embryo patterning and fetal nourishment. While their significance is undeniable, our understanding remains limited, with many questions unanswered about the molecular intricacies driving this development. Revolutionary advancements are emerging, highlighted by the introduction of a groundbreaking synthetic embryoid method and a rapid protein degradation technique. Together, these innovations can reveal the genetic secrets of mammalian fetal development. Our project aims to: 1) Decipher the key transcription factors that shape extraembryonic endoderm development, 2) Understand the signaling molecules that regulate interactions between the embryo and its surrounding extraembryonic tissues, and 3) Construct the first-ever gene regulatory network (GRN) offering a comprehensive blueprint of mammalian extraembryonic endoderm development. Through our efforts, we plan to uncover previously unidentified causes of various congenital defects and pregnancy complications. |